SeptiSearch
a compendium of sepsis pathogenesis genes
SeptiSearch is an effort to aggregate genes from the literature previously implicated in sepsis pathogenesis. While I conceived the idea and managed the project, Jasmine Tam (an undergraduate student at McGill University), performed a literature search of 100+ published transcriptomics studies and curated sepsis-associated genes into a database. Furthermore, Travis Blimike (a staff bioinformatician at the Hancock Lab) built a Shiny app permitting the exploration of these genes (septisearch.ca).
The database is the largest of its kind, and I am currently mining this database for novel biological insights.
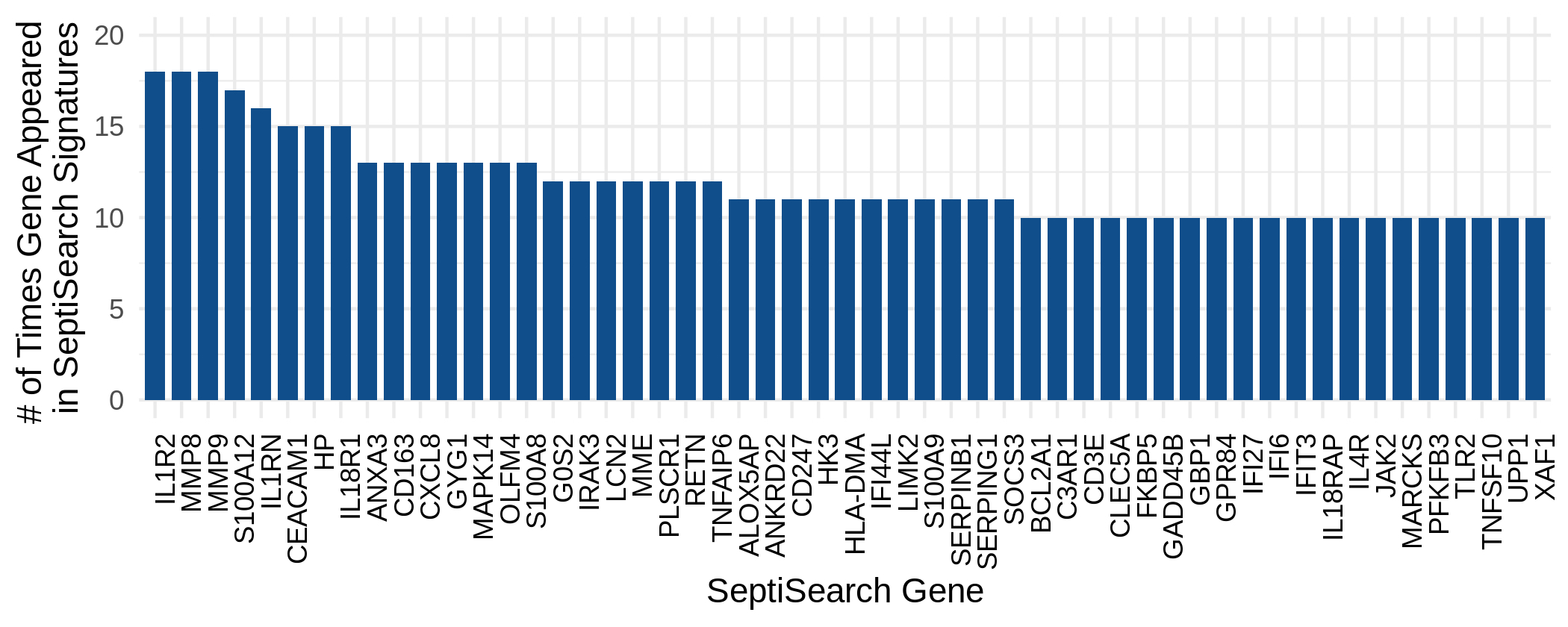
The still growing database comprises 7949 unique genes. This figure shows which genes were described repeatedly across publications, which included Interleukin 1 Receptor Type 2 (IL1R2), Matrix Metallopeptidase 8 and 9 (MMP8/9), S100 Calcium Binding Protein A12 (S100A12), Interleukin 1 receptor antagonist (IL1RN), etc..
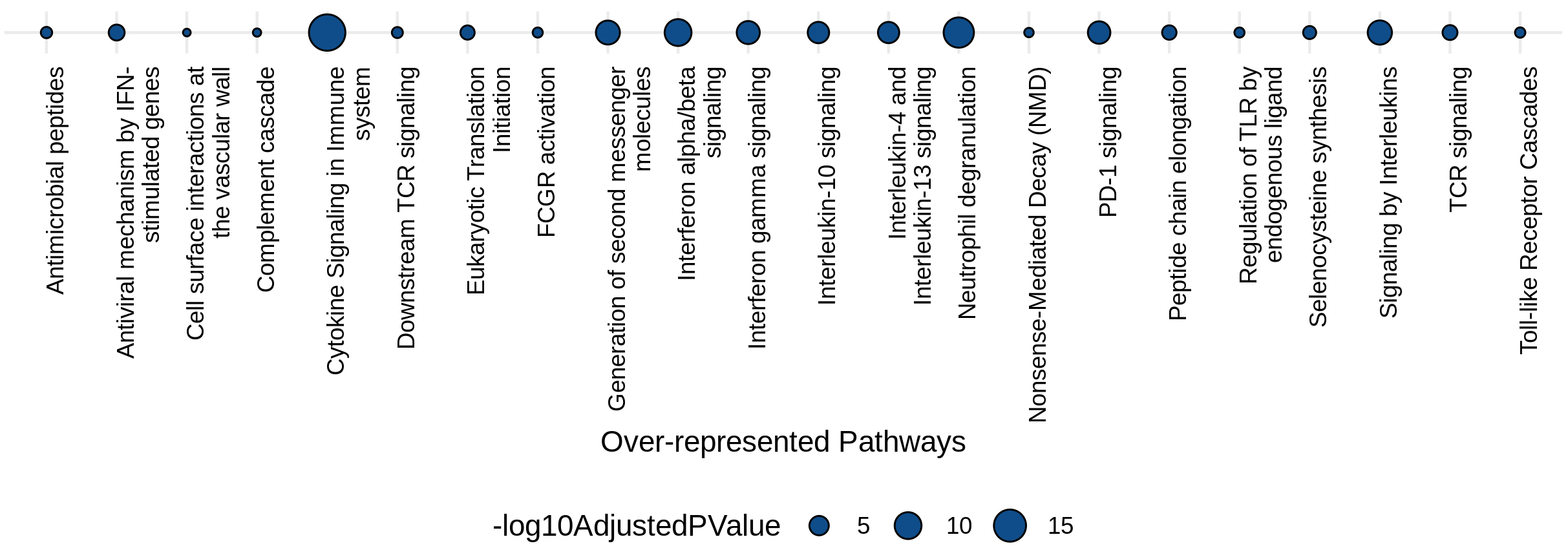
This is a pathway over-representation analysis using the top 200 most frequently identified genes. Significantly over-represented pathways included Neutrophil degranulation, PD-1 signaling, Interferon-alpha/beta signaling, downstream TCR signaling, generation of second messenger molecules, IL-4 and -13 signaling, among others. These pathways are all putative regulators of immune responses. I am preparing a publication with these findings.