Endotypes of early sepsis
a patient subgrouping approach for sepsis diagnosis.
Sepsis patients exhibit a broad spectrum of disease manifestations, ranging from mild to requiring ventilation, organ dysfunction, septic shock, and death. Accordingly, given the clinical variability among sepsis patients, there is considerable difficulty in accurately defining the syndrome, and this is especially true at first clinical presentation (Levy et al. 2021). The issue of heterogeneity has led to sepsis being reframed as a syndrome of distinct endotypes, which are subgroups of patients characterized by distinct pathobiological mechanisms (Leligdowicz et al. 2019).
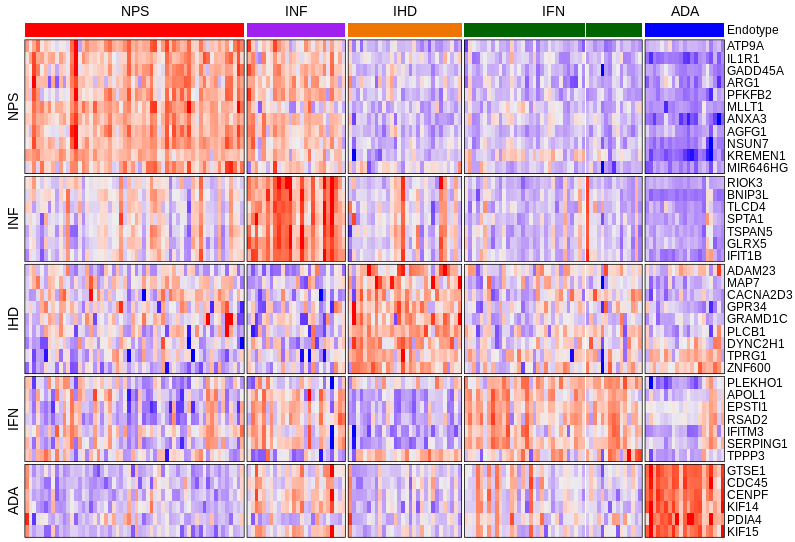
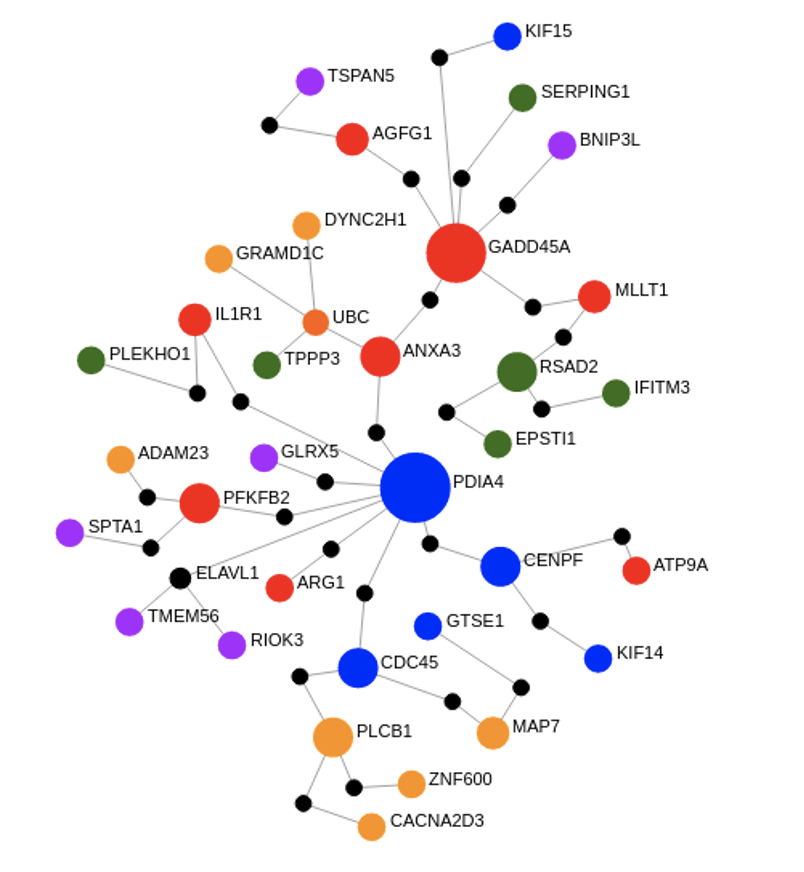
In this project, which also comprised the bulk of my PhD dissertation work, I determined that five molecular gene expression endotypes define the early sepsis syndrome, each having a unique genomic signature (or fingerprint) enabling their detection. Importantly, these endotypes can predict the likelihood a patient will progress to organ dysfunction and septic shock. To identify endotypes, our lab performed whole blood transcriptomics on a total of 260 patients from four global ERs (Netherlands, Colombia, Canada, and Australia).
The study was published in EBioMedicine. We also got quite a bit of press coverage, including stories in CTV News, Medical Xpress, and Vancouver is Awesome.
Techniques used:
- consensus K-medoids clusterings to identify endotypes
- DE and pathway over-representation analysis to characterize endotypes
- training a LASSO-regularized multinomial regression model to enable endotype classification in clinical settings